Explore the data structure of a dynophore
A dynophore (defined in the
Dynophoreclass) is a collection of so-called superfeatures (defined in theSuperFeatureclass). A superfeature is defined as a pharmacophore feature on ligand site (defined by a feature type, e.g. HBA, and one or more ligand atom numbers/serials) that occurs at least once during and MD simulation.A superfeature can have one or more interaction partner(s) on macromolecule-side. These interaction partners are called environmental partners (defined in the
EnvPartnerclass).Each superfeature is described in 3D with a chemical feature point cloud (
ChemicalFeatureCloud3Dclass).Additionally, the dynophore contains information about the bound ligand in the
Ligandclass.
In this notebook, we will explore the Dynophore, Ligand, SuperFeature, EnvPartner, and ChemicalFeatureCloud3D classes.
[1]:
%load_ext autoreload
%autoreload 2
[2]:
from pathlib import Path
import logging
from dynophores import Dynophore
[3]:
logger = logging.getLogger("dynophores")
logger.setLevel(logging.DEBUG)
Set path to DynophoreApp output data folder
[4]:
DATA = Path("../../dynophores/tests/data")
dyno_path = DATA / "out"
Load data as Dynophore object
[5]:
dynophore = Dynophore.from_dir(dyno_path)
List object attributes
Dynophore object attributes
[6]:
dynophore.__dict__
[6]:
{'id': 'dynophore_1KE7',
'ligand': <dynophores.core.ligand.Ligand at 0x7efe50678d90>,
'superfeatures': {'HBA[4618]': <dynophores.core.superfeature.SuperFeature at 0x7efe50673cd0>,
'AR[4605,4607,4603,4606,4604]': <dynophores.core.superfeature.SuperFeature at 0x7efe506202e0>,
'HBD[4598]': <dynophores.core.superfeature.SuperFeature at 0x7efe50620490>,
'HBA[4606]': <dynophores.core.superfeature.SuperFeature at 0x7efe506203d0>,
'AR[4622,4615,4623,4613,4614,4621]': <dynophores.core.superfeature.SuperFeature at 0x7efdee98ce50>,
'HBD[4612]': <dynophores.core.superfeature.SuperFeature at 0x7efe40bd4730>,
'HBA[4619]': <dynophores.core.superfeature.SuperFeature at 0x7efe40bd57f0>,
'HBA[4596]': <dynophores.core.superfeature.SuperFeature at 0x7efe40bd77c0>,
'H[4615,4623,4622,4613,4621,4614]': <dynophores.core.superfeature.SuperFeature at 0x7efe40f19880>,
'H[4599,4602,4601,4608,4609,4600]': <dynophores.core.superfeature.SuperFeature at 0x7efe40efe220>}}
[7]:
dynophore.__dict__.keys()
# NBVAL_CHECK_OUTPUT
[7]:
dict_keys(['id', 'ligand', 'superfeatures'])
A Dynophore object contains:
id: Dynophore identifier (name)ligand: Ligand informationsuperfeatures: Superfeature data (SuperFeatureobjects)
[8]:
print(f"Number of superfeatures: {len(dynophore.superfeatures)}")
# NBVAL_CHECK_OUTPUT
Number of superfeatures: 10
Ligand object attributes
[11]:
dynophore.ligand.__dict__.keys()
# NBVAL_CHECK_OUTPUT
[11]:
dict_keys(['name', 'smiles', 'mdl_mol_buffer', 'atom_serials'])
[12]:
dynophore.ligand.name
# NBVAL_CHECK_OUTPUT
[12]:
'LS3'
[13]:
dynophore.ligand.smiles
# NBVAL_CHECK_OUTPUT
[13]:
'S5(=O)(=O)CC4C(=CC=C(NC=C3C(=O)NC2C3=CC(C1OC=NC=1)=CC=2)C=4)C5'
[15]:
print(dynophore.ligand.atom_serials)
# NBVAL_CHECK_OUTPUT
[4617, 4604, 4618, 4619, 4596, 4606, 4598, 4612, 4605, 4603, 4597, 4599, 4607, 4613, 4602, 4609, 4615, 4621, 4600, 4614, 4623, 4608, 4601, 4622, 4610, 4616, 4620, 4611, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1]
Note: “-1” in case of atoms without serials (e.g. H atoms)
[19]:
dynophore.ligand.rdkit_molecule()
[19]:
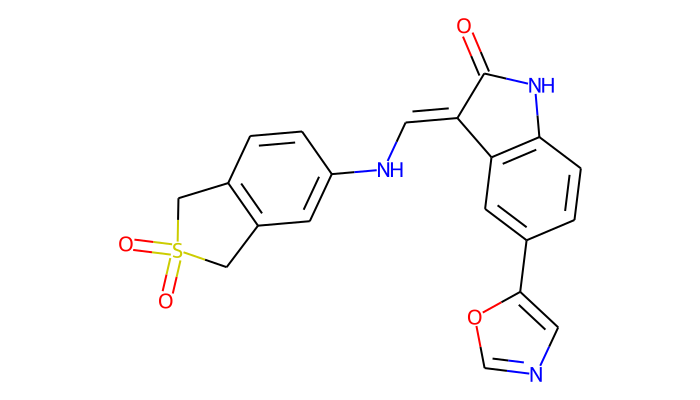
SuperFeature object attributes
Let’s take a look at one example SuperFeature object.
[9]:
superfeature_id = list(dynophore.superfeatures.keys())[0]
superfeature_id
[9]:
'HBA[4618]'
[10]:
dynophore.superfeatures
[10]:
{'HBA[4618]': <dynophores.core.superfeature.SuperFeature at 0x7f050f639f10>,
'AR[4605,4607,4603,4606,4604]': <dynophores.core.superfeature.SuperFeature at 0x7f050f639dc0>,
'HBD[4598]': <dynophores.core.superfeature.SuperFeature at 0x7f055986d040>,
'HBA[4606]': <dynophores.core.superfeature.SuperFeature at 0x7f055986d190>,
'AR[4622,4615,4623,4613,4614,4621]': <dynophores.core.superfeature.SuperFeature at 0x7f055986d640>,
'HBD[4612]': <dynophores.core.superfeature.SuperFeature at 0x7f055986dee0>,
'HBA[4619]': <dynophores.core.superfeature.SuperFeature at 0x7f055986efa0>,
'HBA[4596]': <dynophores.core.superfeature.SuperFeature at 0x7f0559871f70>,
'H[4615,4623,4622,4613,4621,4614]': <dynophores.core.superfeature.SuperFeature at 0x7f0559b72070>,
'H[4599,4602,4601,4608,4609,4600]': <dynophores.core.superfeature.SuperFeature at 0x7f0559b9c9d0>}
[11]:
dynophore.superfeatures[superfeature_id].__dict__.keys()
# NBVAL_CHECK_OUTPUT
[11]:
dict_keys(['id', 'feature_type', 'atom_numbers', 'occurrences', 'envpartners', 'color', 'cloud'])
A SuperFeature object contains:
id: Superfeature identifier (nomenclature:<feature_type><list of atom numbers>)feature_type: Feature type (e.g. HBA, HBD, H, AR, …)atom_numbers: Number(s) of ligand atom(s) that are involved in featureoccurrences: Superfeature occurrences during an MD simulation (0/1 for absent/present)envpartners: Environmental partners on the macromolecule-side that involved in the superfeature (either at the same time or not)cloud: Chemical feature cloud in 3D (coordinates of each occurring feature during an MD simulation that belongs to the superfeature)
[12]:
n_envpartners = sum(
[len(superfeature.envpartners) for _, superfeature in dynophore.superfeatures.items()]
)
print(f"Number of environmental partners: {n_envpartners}")
# NBVAL_CHECK_OUTPUT
Number of environmental partners: 28
EnvPartner object attributes
Let’s take a look at one example EnvPartner object.
[13]:
envpartner_id = list(dynophore.superfeatures[superfeature_id].envpartners.keys())[0]
envpartner_id
[13]:
'LYS-20-A[316]'
[14]:
dynophore.superfeatures[superfeature_id].envpartners[envpartner_id].__dict__.keys()
# NBVAL_CHECK_OUTPUT
[14]:
dict_keys(['id', 'residue_name', 'residue_number', 'chain', 'atom_numbers', 'occurrences', 'distances'])
A EnvPartner object contains:
id: environmental partner identifier (nomenclature:<residue name>-<residue number>-<chain><list of atom numbers>)residue_name: residue nameresidue_number: residue numberchain: chain IDatom_numbers: number(s) of residue atom(s) that are involved in featureoccurrences: interaction occurrences during an MD (0/1 for absent/present) between ligand and residue atomsdistances: interaction distances between the involved atoms on ligand- and macromolecule-side during an MD
ChemicalFeatureCloud3D object attributes
[15]:
dynophore.superfeatures[superfeature_id].cloud.__dict__
[15]:
{'center': array([-18.507637 , -8.405735 , 1.5362723]),
'points': [<dynophores.core.chemicalfeaturecloud3dpoint.ChemicalFeatureCloud3DPoint at 0x7f050f639bb0>,
<dynophores.core.chemicalfeaturecloud3dpoint.ChemicalFeatureCloud3DPoint at 0x7f050f639fd0>]}
[16]:
dynophore.superfeatures[superfeature_id].cloud.__dict__.keys()
# NBVAL_CHECK_OUTPUT
[16]:
dict_keys(['center', 'points'])
A ChemicalFeatureCloud3D object contains:
center: The coordinates of the geometric center of all points in the point cloudpoints: The coordiantes of all points in the point cloud
Dynophore basics
Dynophore identifier
[17]:
print(f"Dynophore name: {dynophore.id}")
Dynophore name: dynophore_1KE7
Number of frames
[18]:
print(f"Number of MD simulation frames: {dynophore.n_frames}")
Number of MD simulation frames: 1002
Number of superfeatures
[19]:
print(f"Number of superfeatures: {dynophore.n_superfeatures}")
Number of superfeatures: 10
Superfeatures monitoring (over trajectory)
Superfeature occurrences
[20]:
dynophore.superfeatures_occurrences
[20]:
| H[4599,4602,4601,4608,4609,4600] | H[4615,4623,4622,4613,4621,4614] | HBA[4596] | HBA[4619] | HBD[4612] | AR[4622,4615,4623,4613,4614,4621] | HBA[4606] | HBD[4598] | HBA[4618] | AR[4605,4607,4603,4606,4604] | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 997 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 998 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 999 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1000 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1001 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
1002 rows × 10 columns
Environmental partners monitoring (over trajectory)
Interaction occurrences for example superfeature
[21]:
dynophore.envpartners_occurrences_by_superfeature(superfeature_id)
[21]:
| LYS-20-A[316] | |
|---|---|
| 0 | 0 |
| 1 | 0 |
| 2 | 0 |
| 3 | 0 |
| 4 | 0 |
| ... | ... |
| 997 | 0 |
| 998 | 0 |
| 999 | 0 |
| 1000 | 0 |
| 1001 | 0 |
1002 rows × 1 columns
[22]:
dynophore.envpartners_occurrences.keys()
[22]:
dict_keys(['HBA[4618]', 'AR[4605,4607,4603,4606,4604]', 'HBD[4598]', 'HBA[4606]', 'AR[4622,4615,4623,4613,4614,4621]', 'HBD[4612]', 'HBA[4619]', 'HBA[4596]', 'H[4615,4623,4622,4613,4621,4614]', 'H[4599,4602,4601,4608,4609,4600]'])
Interaction distances for example superfeature
[23]:
dynophore.envpartners_distances_by_superfeature(superfeature_id)
[23]:
| LYS-20-A[316] | |
|---|---|
| 0 | 11.076880 |
| 1 | 11.076880 |
| 2 | 10.654148 |
| 3 | 9.423526 |
| 4 | 12.006125 |
| ... | ... |
| 997 | 4.339370 |
| 998 | 5.333941 |
| 999 | 5.336269 |
| 1000 | 4.156338 |
| 1001 | 6.207083 |
1002 rows × 1 columns
[24]:
dynophore.envpartners_distances.keys()
[24]:
dict_keys(['HBA[4618]', 'AR[4605,4607,4603,4606,4604]', 'HBD[4598]', 'HBA[4606]', 'AR[4622,4615,4623,4613,4614,4621]', 'HBD[4612]', 'HBA[4619]', 'HBA[4596]', 'H[4615,4623,4622,4613,4621,4614]', 'H[4599,4602,4601,4608,4609,4600]'])
Superfeatures vs. environmental partners
Occurrence count
[25]:
dynophore.count
[25]:
| HBA[4618] | AR[4605,4607,4603,4606,4604] | HBD[4598] | HBA[4606] | AR[4622,4615,4623,4613,4614,4621] | HBD[4612] | HBA[4619] | HBA[4596] | H[4615,4623,4622,4613,4621,4614] | H[4599,4602,4601,4608,4609,4600] | |
|---|---|---|---|---|---|---|---|---|---|---|
| ALA-144-A[2263,2266] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 992 |
| ALA-31-A[488,491] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 216 |
| ASP-86-A[1313] | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ASP-86-A[1319] | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 0 | 0 |
| ASP-86-A[1320] | 0 | 0 | 0 | 0 | 0 | 20 | 0 | 0 | 0 | 0 |
| GLN-131-A[2057] | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GLN-131-A[2061] | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| GLN-131-A[2062] | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| GLU-81-A[1228] | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HIS-84-A[1284,1285,1286,1287,1288] | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ILE-10-A[165] | 0 | 0 | 0 | 0 | 1 | 0 | 116 | 0 | 0 | 0 |
| ILE-10-A[169,171,172] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 995 | 959 |
| ILE-10-A[169,171] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 945 | 972 |
| LEU-134-A[2109,2110,2111] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 999 |
| LEU-83-A[1260] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 862 | 0 | 0 |
| LEU-83-A[1263] | 0 | 0 | 2 | 0 | 0 | 33 | 0 | 0 | 0 | 0 |
| LYS-129-A[2026] | 0 | 1 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 0 |
| LYS-20-A[308] | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| LYS-20-A[316] | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| LYS-89-A[1374] | 0 | 0 | 0 | 0 | 38 | 0 | 0 | 0 | 0 | 0 |
| PHE-82-A[1245,1246,1247,1248,1249,1250] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 0 |
| VAL-18-A[275,276,277] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 955 |
| any | 2 | 1 | 10 | 20 | 40 | 80 | 123 | 862 | 1001 | 1002 |
Occurrence frequency
[26]:
dynophore.frequency
[26]:
| HBA[4618] | AR[4605,4607,4603,4606,4604] | HBD[4598] | HBA[4606] | AR[4622,4615,4623,4613,4614,4621] | HBD[4612] | HBA[4619] | HBA[4596] | H[4615,4623,4622,4613,4621,4614] | H[4599,4602,4601,4608,4609,4600] | |
|---|---|---|---|---|---|---|---|---|---|---|
| ALA-144-A[2263,2266] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 99.00 |
| ALA-31-A[488,491] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 21.56 |
| ASP-86-A[1313] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.00 |
| ASP-86-A[1319] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 1.80 | 0.00 | 0.00 | 0.00 | 0.00 |
| ASP-86-A[1320] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 2.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GLN-131-A[2057] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 |
| GLN-131-A[2061] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.80 | 0.00 | 0.00 | 0.00 | 0.00 |
| GLN-131-A[2062] | 0.0 | 0.0 | 0.0 | 0.2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GLU-81-A[1228] | 0.0 | 0.0 | 0.8 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| HIS-84-A[1284,1285,1286,1287,1288] | 0.0 | 0.0 | 0.0 | 0.0 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ILE-10-A[165] | 0.0 | 0.0 | 0.0 | 0.0 | 0.10 | 0.00 | 11.58 | 0.00 | 0.00 | 0.00 |
| ILE-10-A[169,171,172] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 99.30 | 95.71 |
| ILE-10-A[169,171] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 94.31 | 97.01 |
| LEU-134-A[2109,2110,2111] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 99.70 |
| LEU-83-A[1260] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 86.03 | 0.00 | 0.00 |
| LEU-83-A[1263] | 0.0 | 0.0 | 0.2 | 0.0 | 0.00 | 3.29 | 0.00 | 0.00 | 0.00 | 0.00 |
| LYS-129-A[2026] | 0.0 | 0.1 | 0.0 | 1.8 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| LYS-20-A[308] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 |
| LYS-20-A[316] | 0.2 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.30 | 0.00 | 0.00 | 0.00 |
| LYS-89-A[1374] | 0.0 | 0.0 | 0.0 | 0.0 | 3.79 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| PHE-82-A[1245,1246,1247,1248,1249,1250] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 5.69 | 0.00 |
| VAL-18-A[275,276,277] | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 95.31 |
| any | 0.2 | 0.1 | 1.0 | 2.0 | 3.99 | 7.98 | 12.28 | 86.03 | 99.90 | 100.00 |
Superfeature clouds
[27]:
dynophore.cloud_by_superfeature(superfeature_id)
[27]:
| x | y | z | frame_ix | weight | |
|---|---|---|---|---|---|
| 0 | -18.598375 | -8.370245 | 2.017743 | 959 | 1.0 |
| 1 | -18.416897 | -8.441224 | 1.054801 | 964 | 1.0 |
[28]:
dynophore.clouds.keys()
[28]:
dict_keys(['HBA[4618]', 'AR[4605,4607,4603,4606,4604]', 'HBD[4598]', 'HBA[4606]', 'AR[4622,4615,4623,4613,4614,4621]', 'HBD[4612]', 'HBA[4619]', 'HBA[4596]', 'H[4615,4623,4622,4613,4621,4614]', 'H[4599,4602,4601,4608,4609,4600]'])